Software
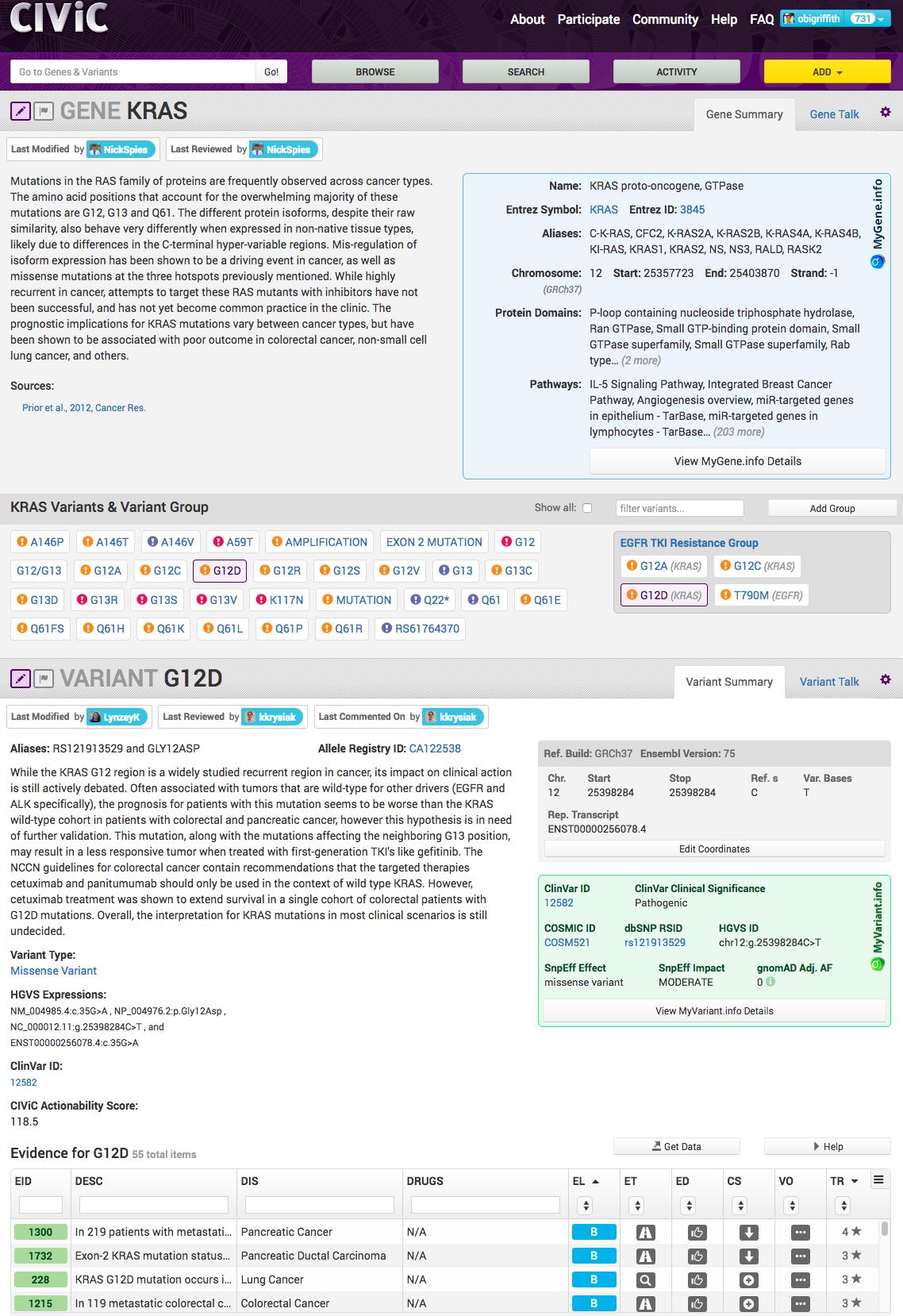

URL: civicdb.org
Description:
CIViC provides a knowledgebase and curation system for expert crowdsourcing the clinical interpretation of variants in cancer.
Team members:
Malachi Griffith, Nicholas Spies, Kilannin Krysiak, Josh McMichael, Adam Coffman, Arpad Danos, Susanna Kiwala, Ben Ainscough, Erica Barnell, Alex Wagner, Zachary Skidmore, Robert Lesurf, Lee Trani, Avinash Ramu, Katie Campbell, Ajay Venigalla, Lana Sheta, Obi Griffith
Source code:
civic-client (GitHub)civic-server (GitHub)
Citation(s):
1. Griffith, Spies, Krysiak et al. 2017. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nature Genetics. 49(2):170–174.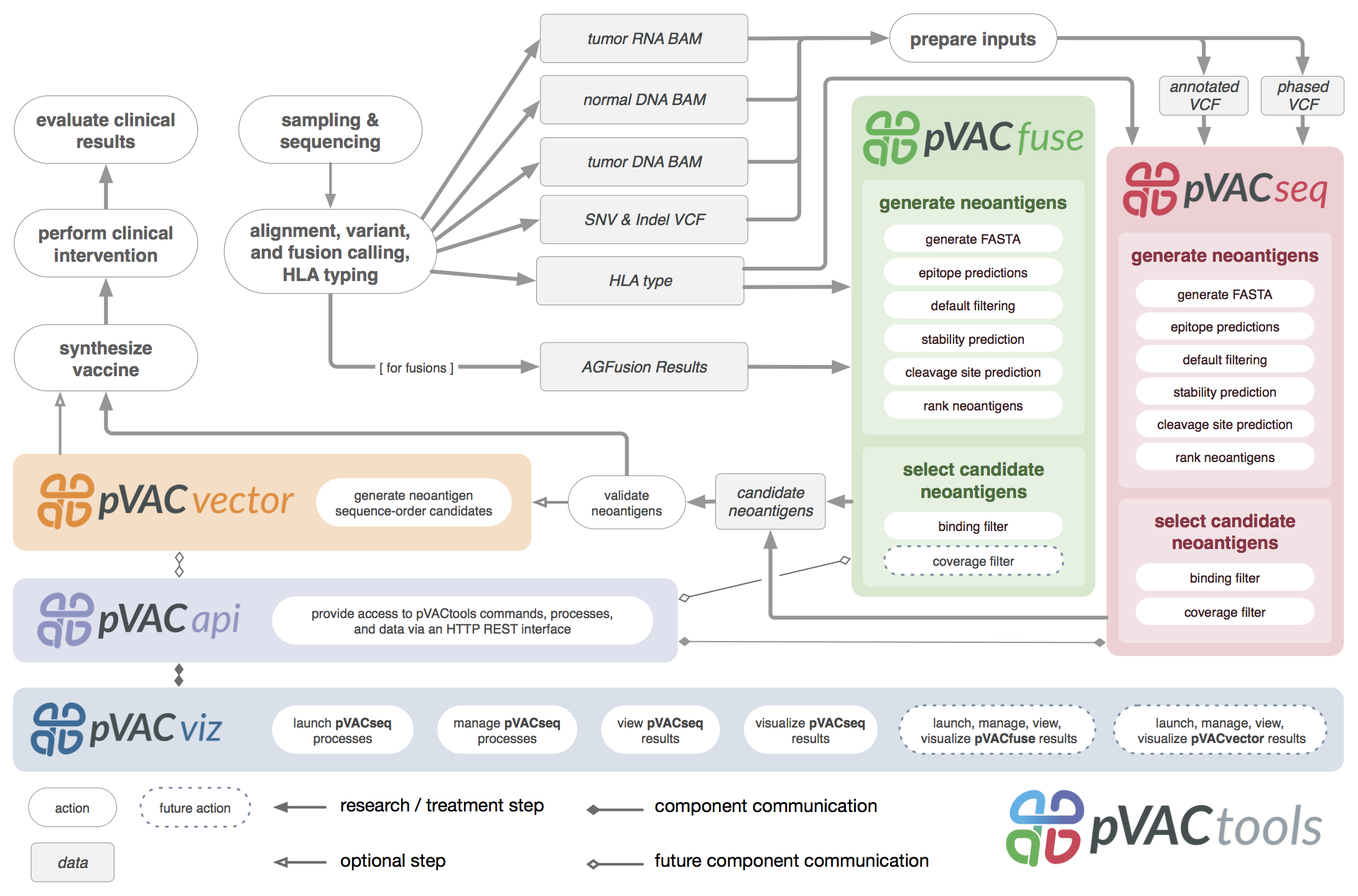

URL: pvactools.org
Description:
A computational toolkit for aiding design of personalized cancer vaccines.
Team members:
Susanna Kiwala, Huiming Xia, Megan Richters, Joshua McMichael, Christopher Miller, Jasreet Hundal, Yang-Yang Feng, Aaron Graubert, Alex Wollam, Amber Wollam, Connor Liu, Jason Walker, Obi Griffith, Malachi Griffith
Source code:
pVACtools (GitHub)Citation(s):
1. Hundal et al. 2016. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Medicine. 8(1):11.2. Hundal, Kiwala et al. 2020. pVACtools: A Computational Toolkit to Identify and Visualize Cancer Neoantigens. Cancer Immunology Research. 8(3):409-420.
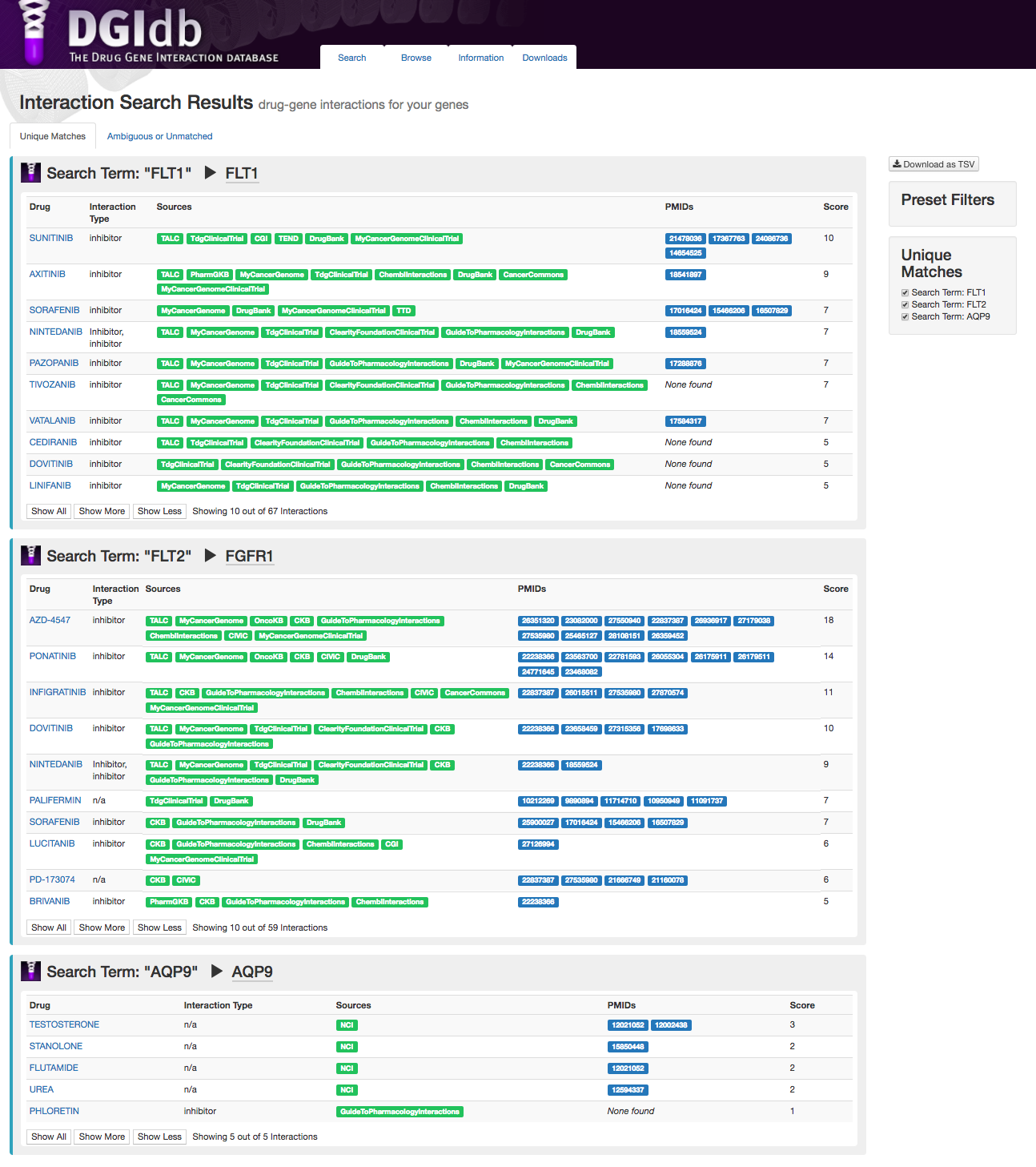

URL: dgidb.org
Description:
DGIdb offers user-friendly browsing, searching, and filtering of information on drug-gene interactions and the druggable genome, mined from over thirty trusted sources.
Team members:
Kelsy Cotto, Alex Wagner, Yang-Yang Feng, Susanna Kiwala, Adam Coffman, Gregory Spies, Alex Wollam, Nicholas Spies, Obi Griffith, Malachi Griffith
Source code:
dgi-db (GitHub)Citation(s):
1. Griffith, Griffith, et al. 2013. DGIdb: mining the druggable genome. Nature Methods. 10(12):1209-10.2. Wagner et al. 2016. DGIdb 2.0: mining clinically relevant drug-gene interactions. Nucleic Acids Research. 44(D1):D1036-44.
3. Cotto, Wagner et al. 2018. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Research.
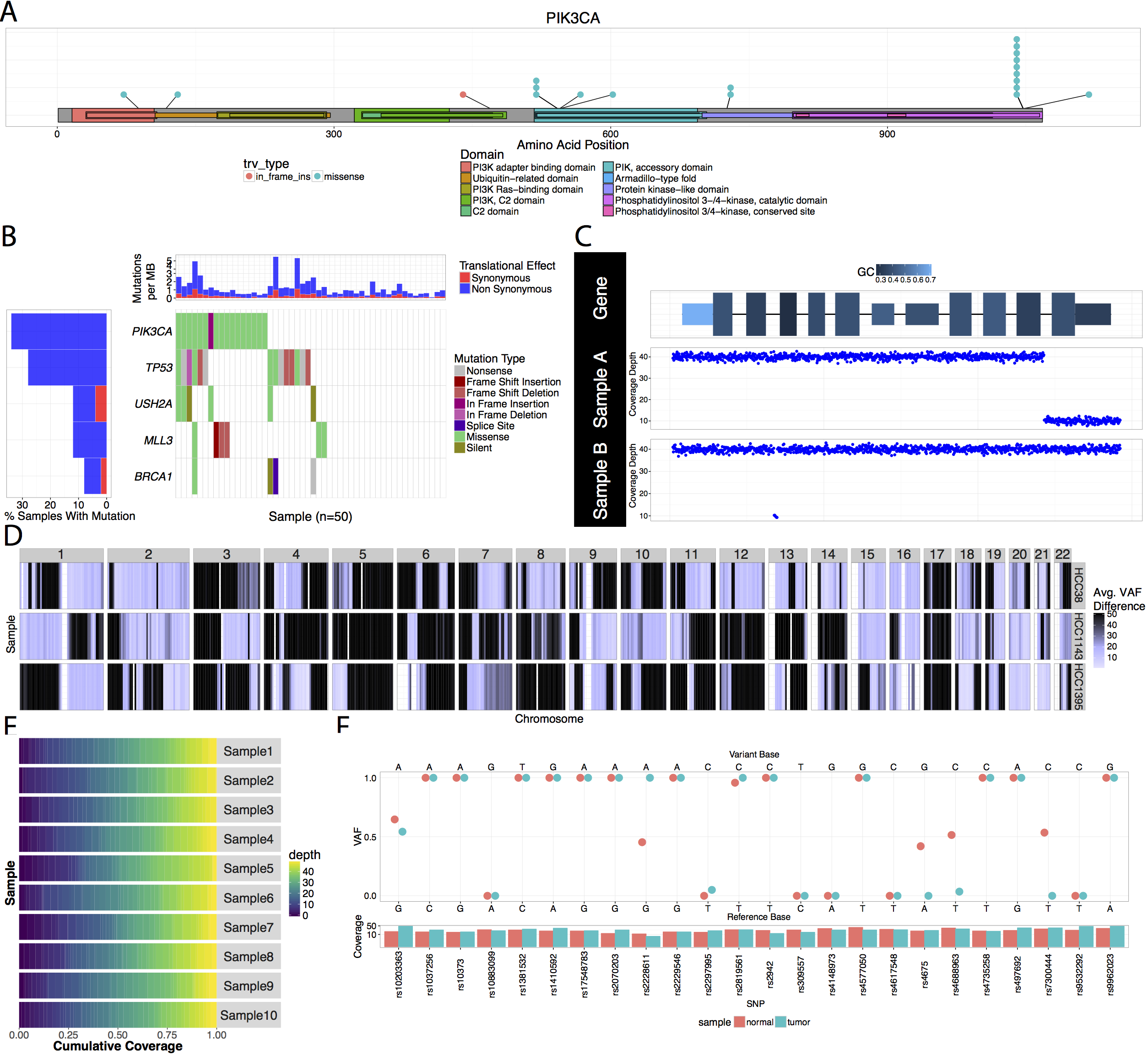
URL: github.com/griffithlab/GenVisR
Description:
GenVisR is a bioconductor package designed to allow users to produce highly customizable publication quality graphics for genomic data primarily at the cohort level.
Team members:
Zachary Skidmore, Alex Wagner, Katie Campbell, Kelsy Cotto, Jason Kunisaki, Obi Griffith, Malachi Griffith.
Source code:
GenVisR (Bioconductor)GenVisR (GitHub)
Citation(s):
1. Skidmore et al. 2016. GenVisR: Genomic Visualizations in R. Bioinformatics. 32(19):3012-4.2. Skidmore et al. 2021. Exploring the Genomic Landscape of Cancer Patient Cohorts with GenVisR. Current Protocols.
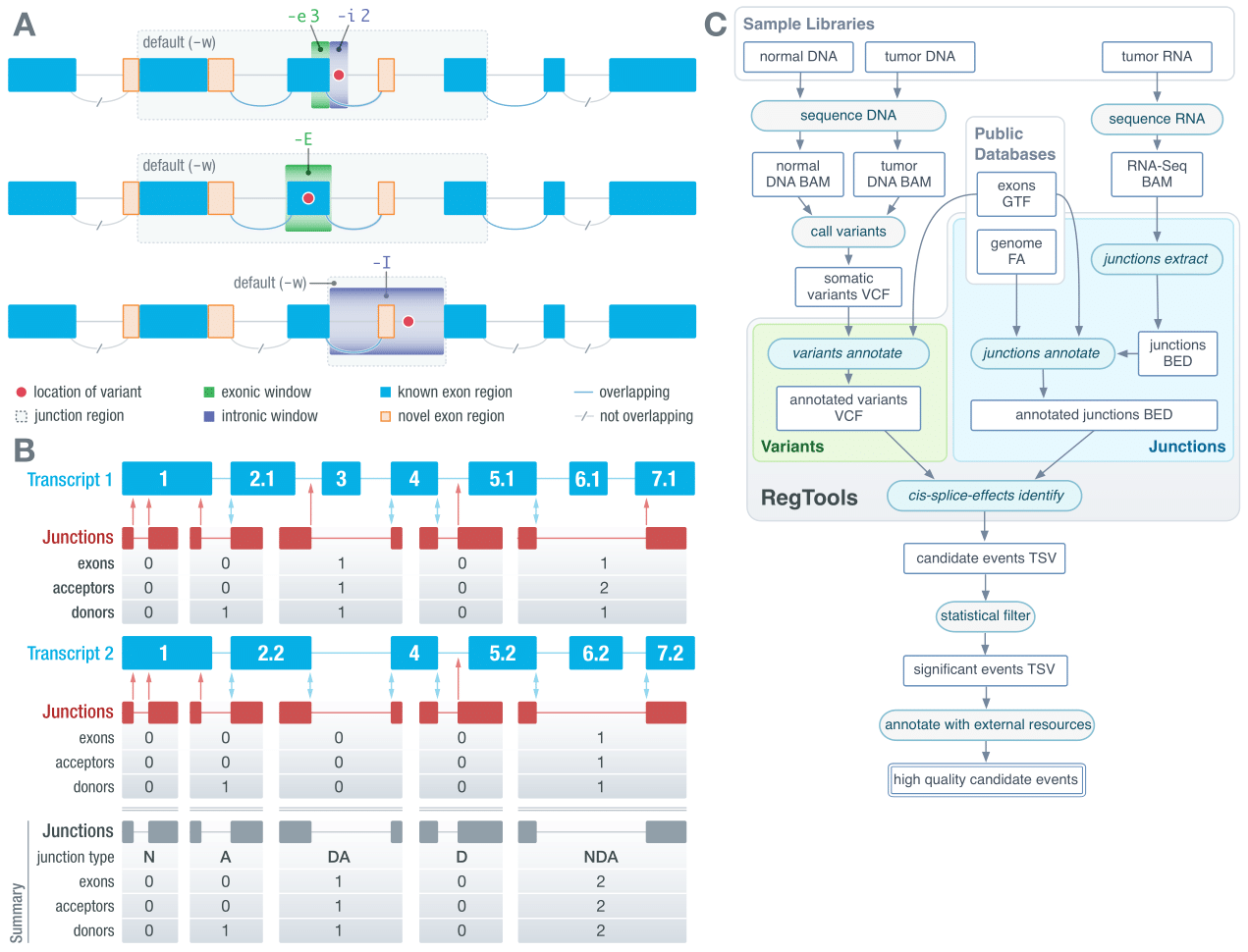
URL: regtools.org
Description:
RegTools is a set of tools that integrate DNA-seq and RNA-seq data to help interpret mutations in a regulatory and splicing context.
Team members:
Yang-Yang Feng, Avinash Ramu, Kelsy Cotto, Jason Kunisaki, Zachary Skidmore, Obi Griffith, Malachi Griffith
Source code:
RegTools (GitHub)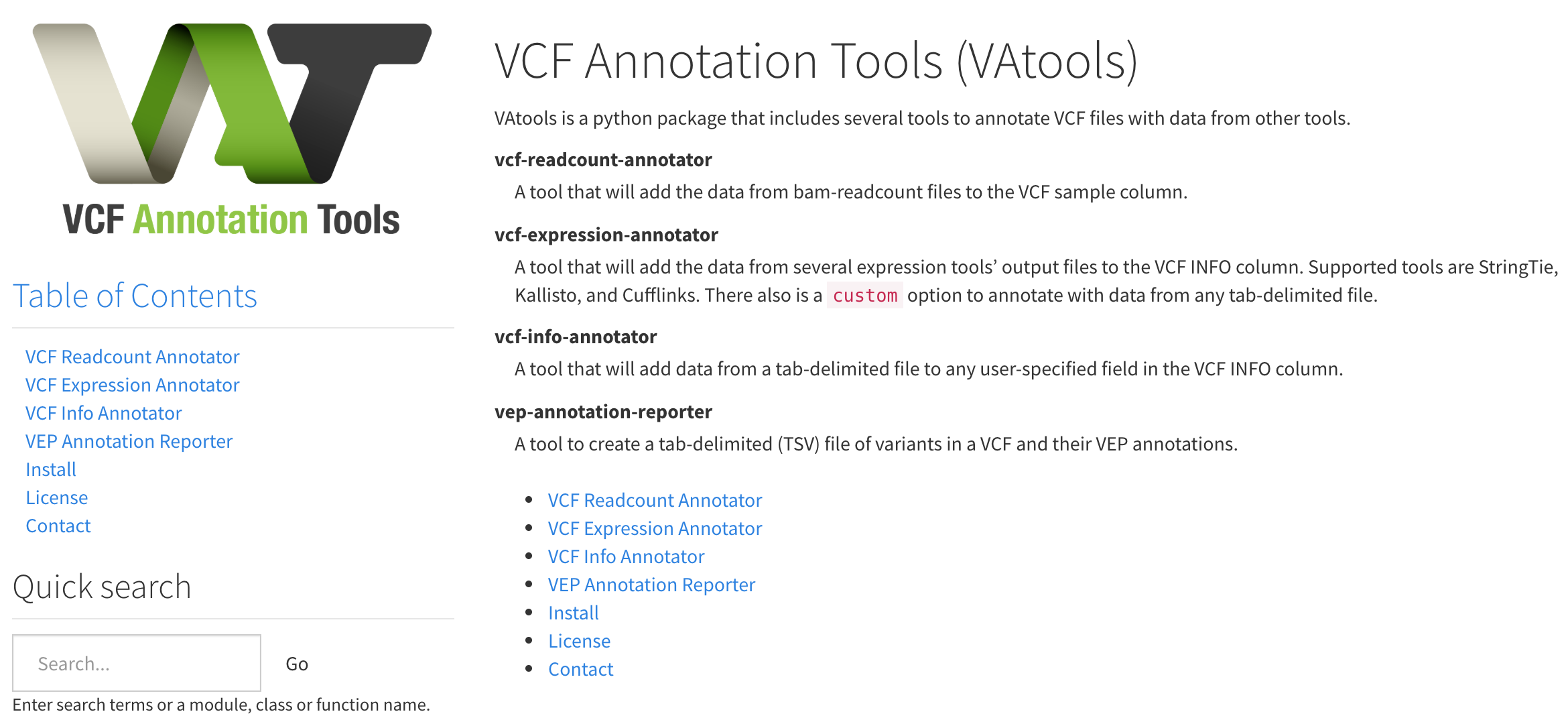

URL: vatools.org
Description:
VAtools is a python package that includes several tools to annotate VCF files with data from other tools.
Team members:
Susanna Kiwala
Source code:
VAtools (GitHub)Citation(s):
1. In preparation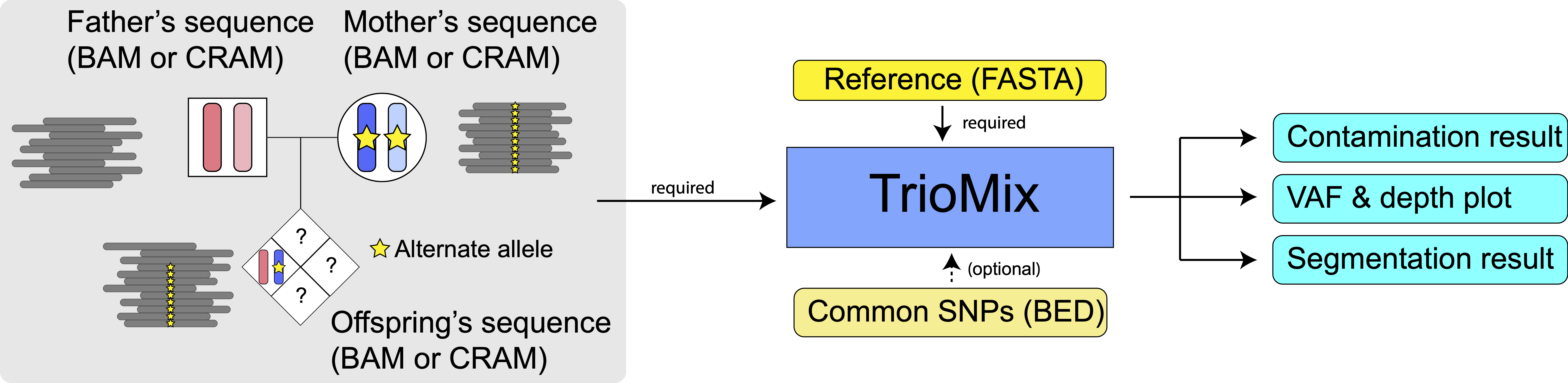
URL: triomix.io
Description:
TrioMix is a software tool for detecting intrafamilial DNA contamination, chimerism, and uniparental disomy from sequencing family trios.
Team members:
Chris Yoon, Obi Griffith, Malachi Griffith
Source code:
TrioMix (GitHub)Citation(s):
1. Yoon et al. 2022. Estimation of intrafamilial DNA contamination in family trio genome sequencing using deviation from Mendelian inheritance. Genome Research (2022).

URL: opencap.org
Description:
An open resource for building custom capture panels that are transparently linked to clinical variant annotations.
Team members:
Erica Barnell, Adam Waalkes, Kelsi Penewit, Katie Campbell, Zachary Skidmore, Colin Pritchard, Todd Fehniger, Ravindra Uppaluri, Ramaswamy Govindan, Malachi Griffith, Stephen Salipante, Obi Griffith
Source code:
OpenCAP (GitHub)Citation(s):
1. Barnell et al. 2019. Open-Sourced CIViC Annotation Pipeline to Identify and Annotate Clinically Relevant Variants Using Single-Molecule Molecular Inversion Probes. JCO Clin Cancer Inform. 3:1-12.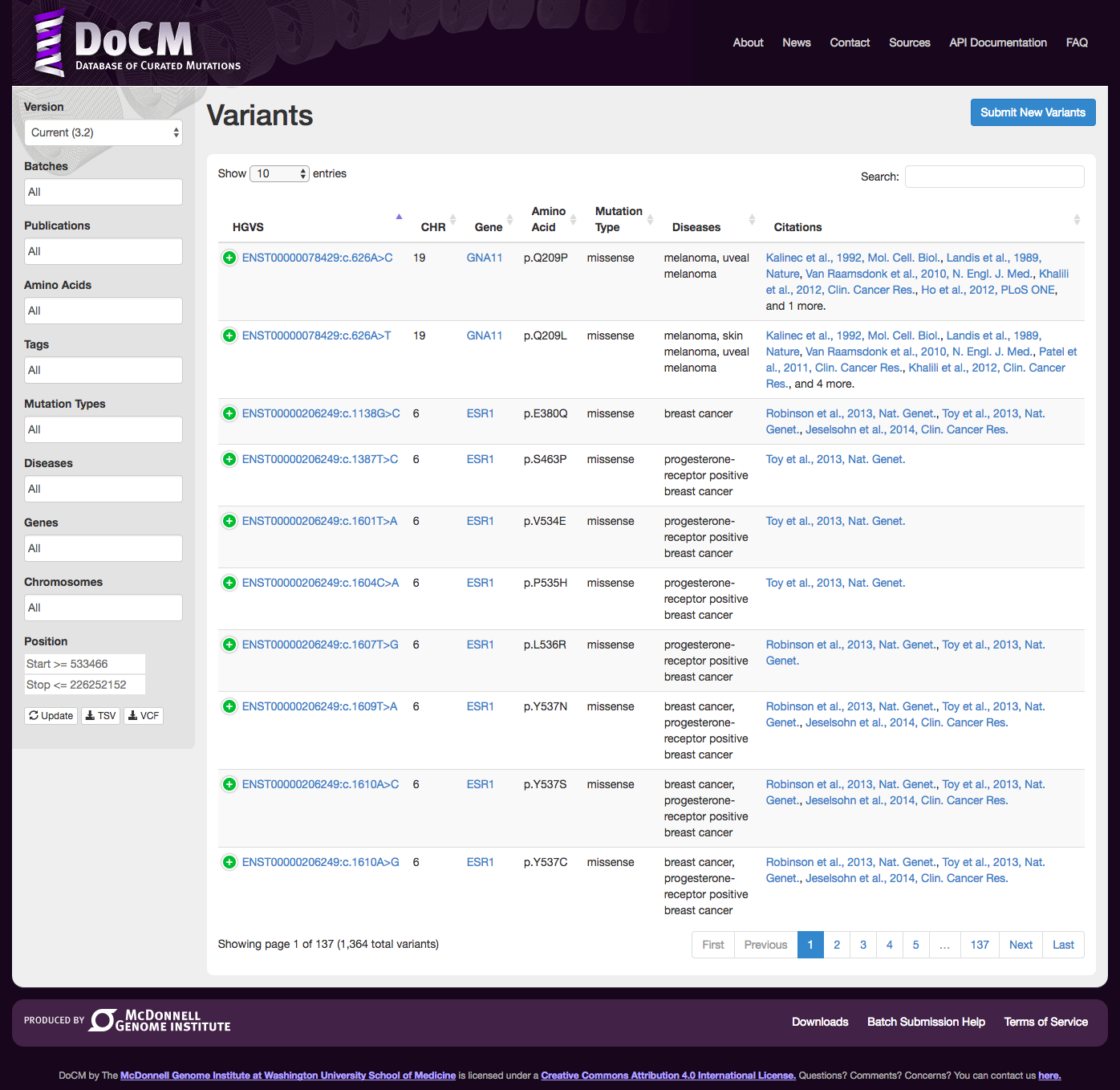

URL: docm.info
Description:
The Database of Curated Mutations (DoCM) is an open access resource that outlines literature supported somatic mutations that are clinically or biologically relevant.
Team members:
Ben Ainscough, Malachi Griffith, Adam Coffman, Alex Wagner, Jason Kunisaki, Joshua McMichael, Obi Griffith
Source code:
docm (GitHub)Citation(s):
1. Ainscough et al. 2016. DoCM: a database of curated mutations in cancer. Nature Methods. 13(10):806-7.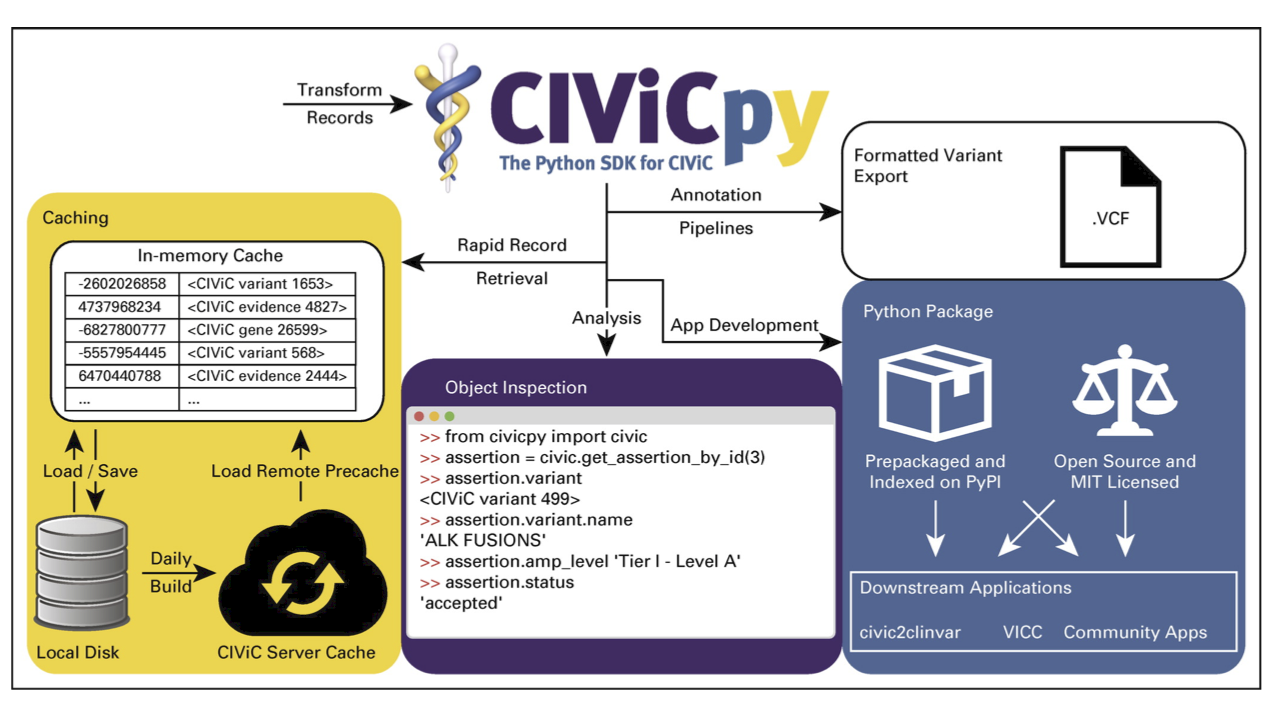

URL: civicpy.org
Description:
CIViCpy is designed to be an intuitive SDK and analysis toolkit for the Clinical Interpretations of Variants in Cancer (CIViC) knowledgebase.
Team members:
Alex Wagner, Susanna Kiwala, Adam Coffman, Joshua McMichael, Kelsy Cotto, Thomas Mooney, Erica Barnell, Kilannin Krysiak, Arpad Danos, Jason Walker, Obi Griffith, Malachi Griffith
